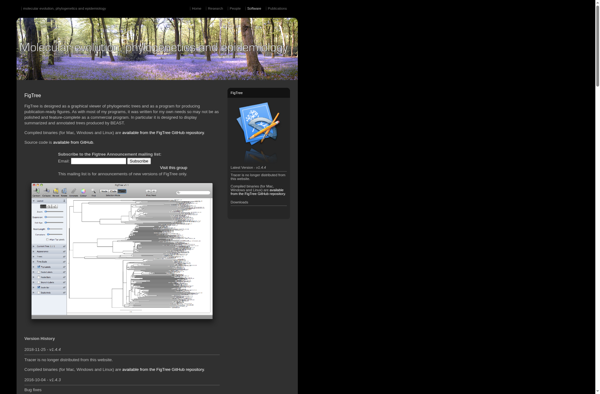
Description: FigTree is a free, open-source desktop application for drawing and viewing phylogenetic trees. It allows easy editing of basic tree aesthetics, such as node shape, color, size, and line width. It can export trees in a range of file formats for publishing and supports large datasets.
Type: Open Source Test Automation Framework
Founded: 2011
Primary Use: Mobile app testing automation
Supported Platforms: iOS, Android, Windows
Description: Archaeopteryx is an open-source genome browser and annotation tool that allows users to visualize, manage, and analyze genomic datasets. It supports a range of common genomics file formats and provides features like dynamic zooming, metadata editing, and sharing sessions.
Type: Cloud-based Test Automation Platform
Founded: 2015
Primary Use: Web, mobile, and API testing
Supported Platforms: Web, iOS, Android, API