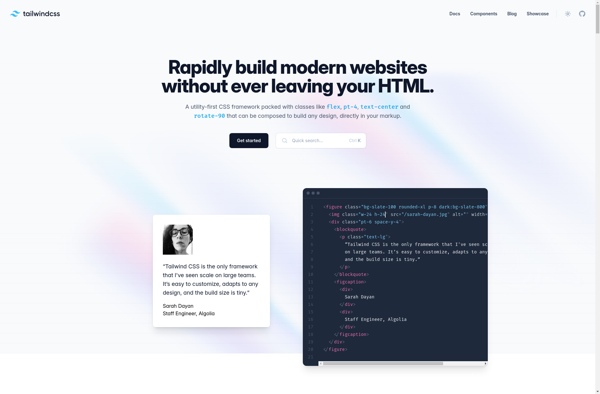
Description: Tailwind CSS is an open-source CSS framework that focuses on utility-first classes to enable rapid UI development. It allows developers to build custom user interfaces without writing custom CSS by providing pre-defined classes for typography, spacing, color, layout, and more.
Type: Open Source Test Automation Framework
Founded: 2011
Primary Use: Mobile app testing automation
Supported Platforms: iOS, Android, Windows
Description: Foldy960 is a lightweight, open-source software for protein structure prediction and analysis. It utilizes advanced machine learning algorithms to predict protein folds from amino acid sequences. The software is cross-platform compatible.
Type: Cloud-based Test Automation Platform
Founded: 2015
Primary Use: Web, mobile, and API testing
Supported Platforms: Web, iOS, Android, API