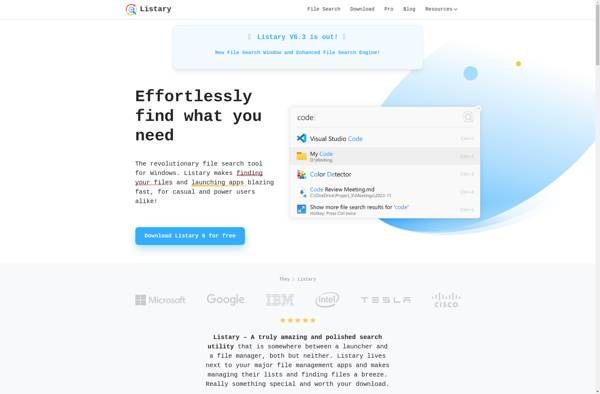
Description: Listary is a file browser enhancement and file launcher program for Windows. It helps organize and access files and folders faster with features like fuzzy search, hotkeys, bookmarks, and virtual folders. Listary increases productivity for power users by making file navigation and opening extremely fast.
Type: Open Source Test Automation Framework
Founded: 2011
Primary Use: Mobile app testing automation
Supported Platforms: iOS, Android, Windows
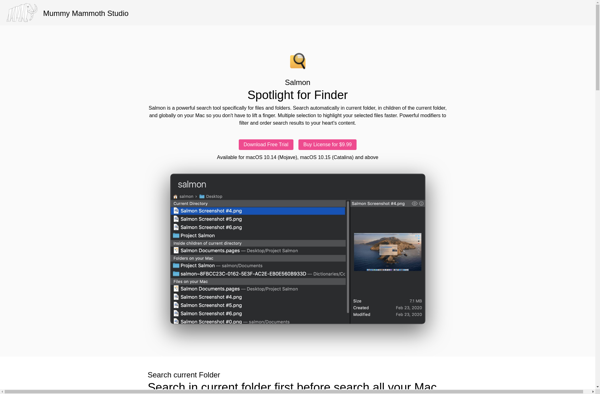
Description: Salmon is an open-source software tool for estimating transcript abundance from RNA-seq data. It uses a model-based approach to align RNA-seq reads to a reference transcriptome and quantify abundance at the transcript level.
Type: Cloud-based Test Automation Platform
Founded: 2015
Primary Use: Web, mobile, and API testing
Supported Platforms: Web, iOS, Android, API