Salmon
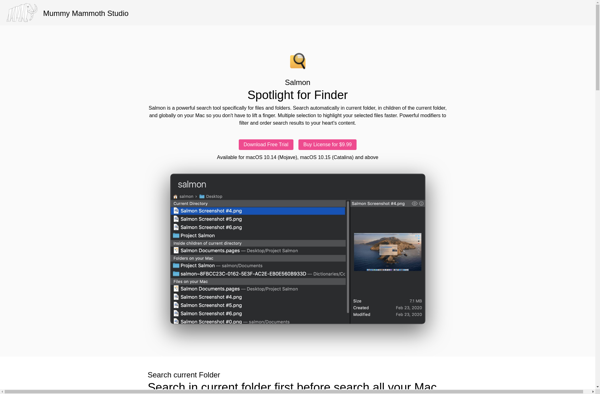
Salmon: Open-Source RNA-Seq Analysis Tool
Estimate transcript abundance from RNA-seq data with Salmon, an open-source software tool that uses a model-based approach for accurate alignment and quantification.
What is Salmon?
Salmon is an open-source software tool developed primarily at the University of California, Berkeley for estimating transcript abundance from RNA-sequencing (RNA-seq) data. It employs a lightweight mapping model to align RNA-seq reads to a reference transcriptome and uses maximum likelihood estimation to quantify abundance at the transcript level.
Some key features and advantages of Salmon include:
- Very fast processing speeds by avoiding alignment of individual bases
- Model-based approach for accurate abundance estimates even for fragmented transcripts
- Can work directly with FASTA/Q files as input
- Provides both quant.sf and quant.genes files summarizing abundance
- Actively maintained and supported on GitHub
Salmon continues to be improved and expanded with additional features for handling various types of RNA-seq data. Its speed, accuracy and lightweight implementation have made it a popular choice for quantifying transcript abundance, especially for large complex transcriptomes.
Salmon Features
Features
- Alignment of RNA-seq reads to a reference transcriptome
- Quantification of transcript abundance
- Support for single-end and paired-end reads
- Bias modeling and correction
- Multi-mapping reads handling
- GC content bias correction
- Strand-specific protocols
- Bootstrapping for confidence interval estimation
- Parallel processing support
Pricing
- Open Source
Pros
Cons
Official Links
Reviews & Ratings
Login to ReviewThe Best Salmon Alternatives
Top Science & Education and Bioinformatics and other similar apps like Salmon
Here are some alternatives to Salmon:
Suggest an alternative ❐Alfred
Raycast
Quicksilver
Ulauncher
Albert
Listary
Gnome Do
Wox
Keypirinha
Cerebro
Fluent Search