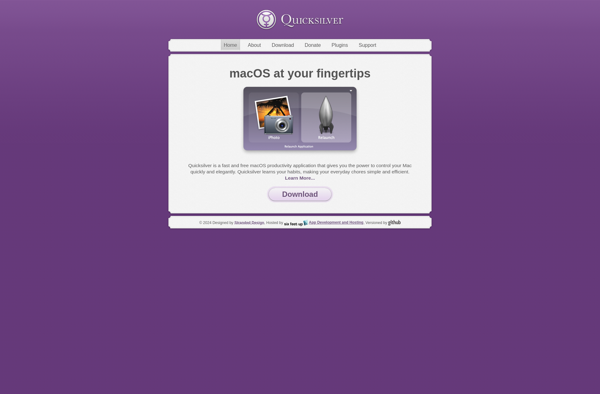
Description: Quicksilver is a free and open-source utility application for macOS that provides quick access to files, contacts, applications and other system tools through an intuitive interface. It allows searching and launching apps and files quickly using abbreviations and keywords.
Type: Open Source Test Automation Framework
Founded: 2011
Primary Use: Mobile app testing automation
Supported Platforms: iOS, Android, Windows
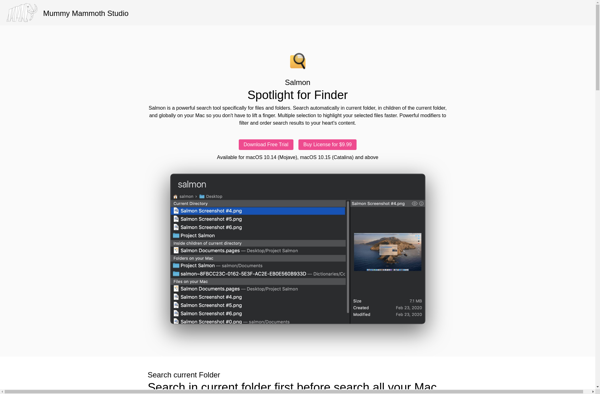
Description: Salmon is an open-source software tool for estimating transcript abundance from RNA-seq data. It uses a model-based approach to align RNA-seq reads to a reference transcriptome and quantify abundance at the transcript level.
Type: Cloud-based Test Automation Platform
Founded: 2015
Primary Use: Web, mobile, and API testing
Supported Platforms: Web, iOS, Android, API