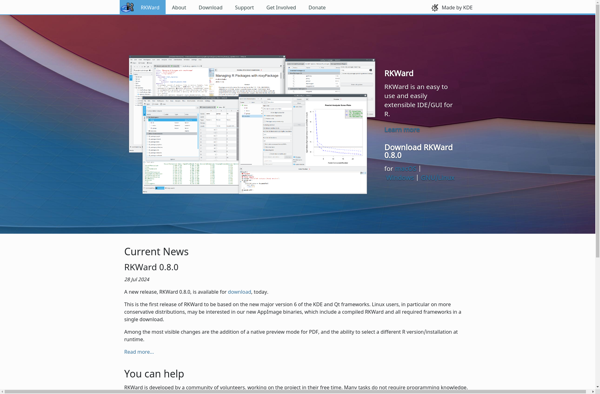
Description: RKWard is an open-source graphical user interface for the R statistical programming language. It provides an integrated development environment to work with R code, data, plots, models and reports.
Type: Open Source Test Automation Framework
Founded: 2011
Primary Use: Mobile app testing automation
Supported Platforms: iOS, Android, Windows
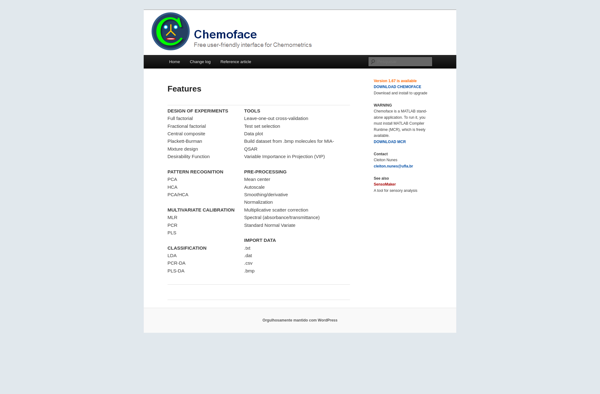
Description: Chemoface is open-source software for predicting the biological activities of small molecules based on their chemical structures. It uses machine learning models trained on datasets of compounds and their bioactivities.
Type: Cloud-based Test Automation Platform
Founded: 2015
Primary Use: Web, mobile, and API testing
Supported Platforms: Web, iOS, Android, API