Chemoface
Chemoface: Open-Source Software for Predicting Biological Activities
Open-source software for predicting biological activities of small molecules based on chemical structures, using machine learning models trained on compound datasets.
What is Chemoface?
Chemoface is an open-source computer program for predicting the biological activities of chemical compounds. It utilizes machine learning models that have been trained on large datasets of chemicals and their associated bioassay data to predict potential therapeutic effects and safety risks.
The key capabilities of Chemoface include:
- Predicting activity against a range of drug targets like enzymes, ion channels, and protein-protein interactions
- Estimating absorption, distribution, metabolism, excretion, and toxicity (ADMET) parameters
- Screening libraries of compounds to identify structures with desired bioactivities
- Prioritizing compounds for synthesis and testing
- Understanding structure-activity relationships
Some of the machine learning methods used by Chemoface include neural networks, random forests, support vector machines, and graph convolutions. It can make predictions based on simplified molecular-input line-entry system (SMILES) representations of compound structures.
Chemoface is implemented in Python and has an open-source MIT license allowing free usage. It provides both command line and graphical user interfaces. The models are customizable and new training data can be added to tailor predictions to specific drug discovery projects.
Chemoface Features
Features
- Predict biological activities of small molecules
- Uses machine learning models trained on bioactivity datasets
- Open-source software
- Web-based graphical user interface
- Support for multiple machine learning algorithms
- Built-in datasets of compounds and bioactivities
- Custom model training
- Activity predictions and statistical analysis
- 2D and 3D molecular structure visualization
- Structure-based virtual screening
Pricing
- Open Source
Pros
Cons
Official Links
Reviews & Ratings
Login to ReviewThe Best Chemoface Alternatives
Top Ai Tools & Services and Machine Learning and other similar apps like Chemoface
Here are some alternatives to Chemoface:
Suggest an alternative ❐R (programming language)
Minitab
RStudio
Deducer
RKWard
DOE++
PSPP
SOFA Statistics
SAS JMP
The R Commander
Statgraphics Centurion XVII
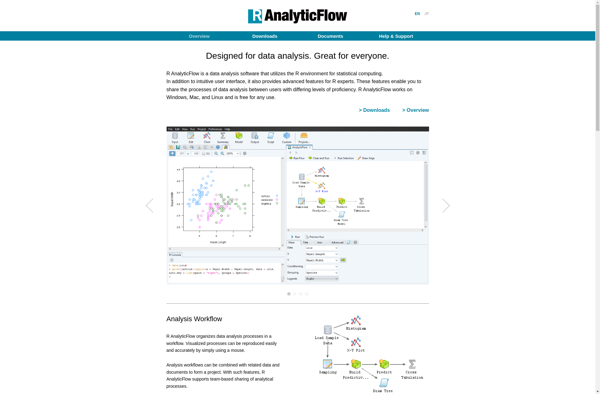
R AnalyticFlow
PAST - PAlaeontological STatistics
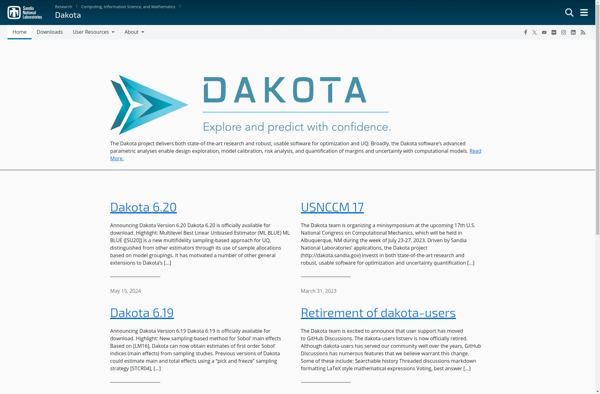
Dakota
Revolution R
SOSstat
