OpenCFU
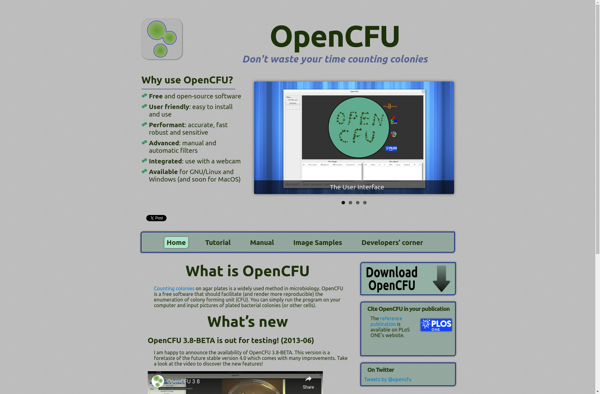
OpenCFU: Open-Source Cell Colony Counter Software
Open-source software for accurate cell colony counting, offering image segmentation and analysis tools for bacterial colonies on agar plates or microscope slides.
What is OpenCFU?
OpenCFU is an open-source, cross-platform software application for counting cell colonies from digital images of agar plates or microscope slides. It provides accurate, automated quantification and analysis of colonies formed by bacteria, yeast, or mammalian cells.
Key features of OpenCFU include:
- Image processing algorithms for identifying, segmenting and quantifying cell colonies
- Colony measurement tools for statistics like colony area, eccentricity, etc.
- Flexible workflow supporting counting of multiple plate images in batches
- Customizable detection settings to optimize counting accuracy
- Data and image export options for further analysis
- Cross-platform support for Windows, Mac and Linux
The intuitive graphical interface allows configuring detection parameters like minimum/maximum colony size to minimize errors. Batch analysis of large sets of plate images is supported for high throughput needs. Quantitative data can be exported as spreadsheet along with colony overlay images showing counting performance.
OpenCFU facilitates reproducible and precise quantification of cell culture assays in microbiology and cell biology research experiments. Its accuracy compares well to manual counting but with much higher throughput.
OpenCFU Features
Features
- Image segmentation and analysis
- Colony counting
- Plate mapping
- Data analysis and statistics
- Customizable workflows
- Open-source and cross-platform
Pricing
- Open Source
Pros
Cons
Official Links
Reviews & Ratings
Login to ReviewThe Best OpenCFU Alternatives
Top Science & Education and Biology & Chemistry and other similar apps like OpenCFU
Here are some alternatives to OpenCFU:
Suggest an alternative ❐Promega Colony Counter
ColonyCount
Colony Counter ( automated )
Colony Counter (manual count)