Bowtie
Bowtie: Ultrafast DNA Sequencing Read Aligner
An ultrafast, memory-efficient short read aligner for aligning short DNA sequencing reads to large genomes, optimized for Illumina, Ion Torrent, and SOLiD sequencing platforms.
What is Bowtie?
Bowtie is an open-source tool for aligning short DNA sequencing reads to large genomes. It was developed at Johns Hopkins University and is optimized for speed, memory efficiency, and sensitivity. Some key features of Bowtie include:
- Extremely fast performance - can align 35-basepair reads to the human genome at a rate of over 25 million reads per CPU hour
- Memory efficient - uses a quality-aware, greedy, randomized algorithm to align reads, minimizing memory footprint
- Supports gapped, local, and paired-end alignment modes
- Outputs alignments in SAM format, allowing interoperation with downstream tools like SAMtools
- Allows mismatches and supports configurable mismatch penalties
- Supports both basespace and colorspace (SOLiD) read alignments
Bowtie is implemented in C++ and is commonly used for a variety of RNAseq, ChIP-seq, and DNA-seq experiments that utilize high-throughput sequencing technologies from Illumina, Ion Torrent, SOLiD, and others. It continues to be maintained and updated to handle newer read lengths and sequencing platforms.
Bowtie Features
Features
- Ultrafast alignment of short DNA sequencing reads
- Memory-efficient - low RAM usage
- Supports aligning reads from Illumina, Ion Torrent, SOLiD platforms
- Supports gapped, local and end-to-end alignments
- Output in SAM/BAM format
Pricing
- Open Source
Pros
Cons
Reviews & Ratings
Login to ReviewThe Best Bowtie Alternatives
Top Science & Education and Bioinformatics and other similar apps like Bowtie
Here are some alternatives to Bowtie:
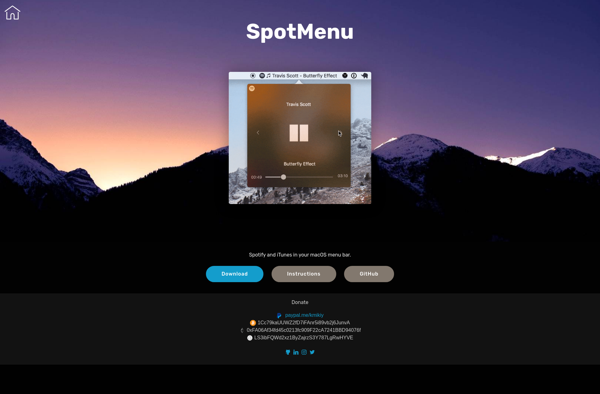
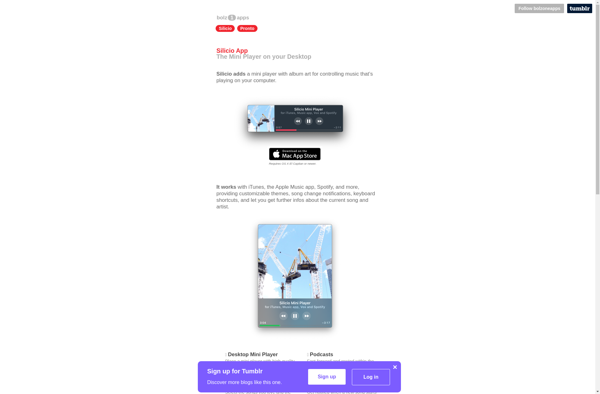
Suggest an alternative ❐Silicio Mini Player
Ecoute
Spotify Now Playing
BarTunes
TunesArt
Gimme Some Tune
NepTunes
CoverSutra