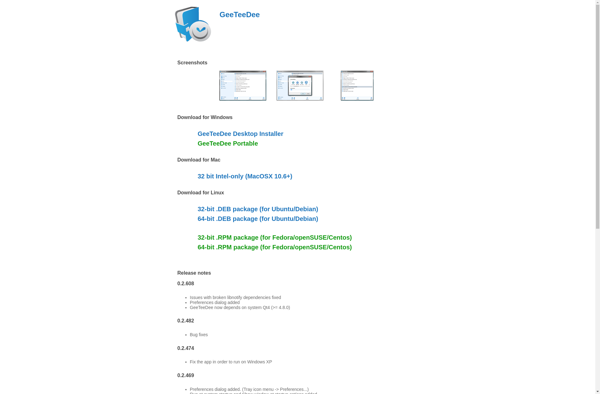
MGSD
mGSD: Open-Source Molecular Dynamics Simulation Software
A cross-platform molecular dynamics simulation software package providing tools for building, running, and analyzing simulations, aiming to be user-friendly and accessible for non-experts.
What is MGSD?
mGSD is an open-source, cross-platform molecular dynamics simulation software package developed by Academic University. It provides advanced tools for building molecular systems, setting up and running simulations, and analyzing the results.
Some key features of mGSD include:
- An intuitive graphical user interface for setting up molecular systems
- Support for a wide range of force fields and simulation parameters
- Built-in scripts and workflows for common simulation tasks
- Scalable simulations that can leverage high-performance computing resources
- Powerful analysis tools including trajectory visualization, clustering, free energy calculations, and more
mGSD aims to make molecular simulations more accessible to both experts and non-experts in the field. Its emphasis on usability and documentation allows users to be productive quickly. The open-source code base encourages customization and collaboration within the user community.
If you are looking for an advanced, flexible, and user-friendly platform for running and analyzing molecular dynamics simulations, mGSD is worth considering.
MGSD Features
Features
- Molecular dynamics simulations
- Force fields for proteins, lipids, nucleic acids
- Graphical user interface
- Parallelization with MPI
- Analysis tools
Pricing
- Open Source
Pros
Cons
Official Links
Reviews & Ratings
Login to ReviewThe Best MGSD Alternatives
Top Science & Engineering and Molecular Modeling and other similar apps like MGSD
Here are some alternatives to MGSD:
Suggest an alternative ❐Todoist
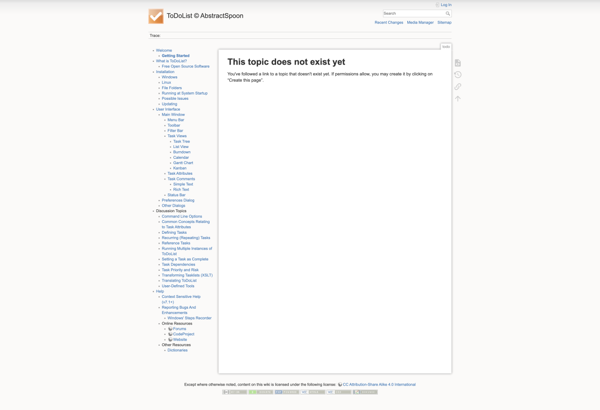
ToDoList
EssentialPIM
MyLifeOrganized
Task Coach
Week Plan

Lifestyle Inspector
Actions by Moleskine

GTDNext
Hero Panel
Wieldy
GeeTeeDee