UCSF Chimera
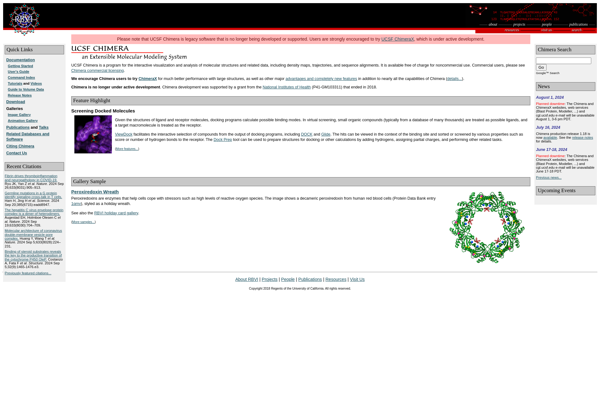
UCSF Chimera: Interactive Molecular Modeling
Extensible molecular modeling and visualization program for interactive analysis and rendering of molecular structures and related data
What is UCSF Chimera?
UCSF Chimera is a highly extensible program for interactive visualization and analysis of molecular structures and related data, including density maps, supramolecular assemblies, sequence alignments, docking results, trajectories, and conformational ensembles. High-quality images and animations can be generated.
Chimera includes complete documentation and several tutorials. Core attributes include:
- highly integrated tools for interactive visualization
- fast rendering and rotatable images even for very large structures
- combination of structure data from PDB files, various types of density maps, sequence alignments, docking results, molecular dynamics trajectories, and other sources
- intuitive and consistent user interface shared among all components
- easily extensible through the customization and development of new tools with the included SDK
Intended uses of UCSF Chimera include examining protein-protein and protein-nucleic acid interactions, assembly of macromolecular complexes, virus particle glycoprotein spikes, organelle membranes, microtubules, and much more. It can also generate high-quality renderings for figures in publications.
UCSF Chimera Features
Features
- Interactive molecular visualization
- High-quality rendering
- Analysis tools for conformations, surfaces, volumes, sequences, docking
- Extensibility through Python scripting
- Support for VR headsets
- Multi-scale modeling
- Integrates with online resources like PDB and PubChem
Pricing
- Free
- Open Source
Pros
Cons
Official Links
Reviews & Ratings
Login to ReviewThe Best UCSF Chimera Alternatives
Top Science & Education and Molecular Modeling and other similar apps like UCSF Chimera
Here are some alternatives to UCSF Chimera:
Suggest an alternative ❐PyMOL
Rasmol
Jmol
VMD - Visual Molecular Dynamics
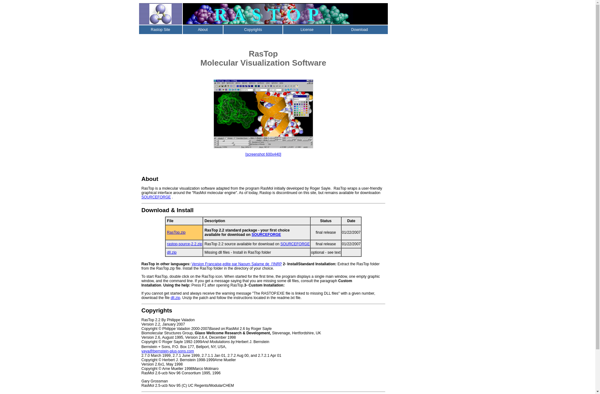
RasTop