VMD - Visual Molecular Dynamics
VMD: Visual Molecular Dynamics
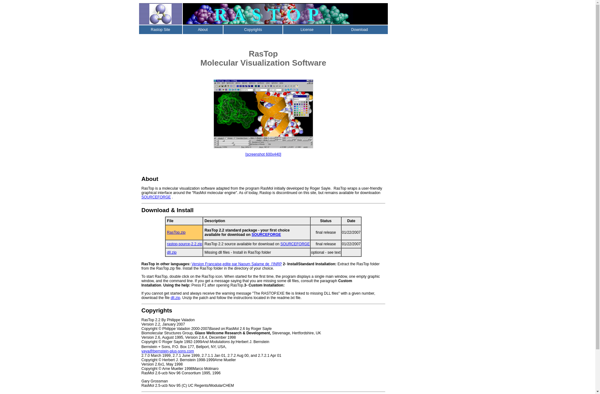
VMD is an open-source molecular visualization program used to visualize, analyze, and animate biological systems such as proteins, nucleic acids, lipid bilayer assemblies. It can handle systems with millions of atoms.
What is VMD - Visual Molecular Dynamics?
VMD (Visual Molecular Dynamics) is a molecular visualization program designed for the display, animation, and analysis of large biomolecular systems using 3-D graphics and built-in scripting. VMD can read standard protein databank files and display the contained structure's secondary structure cartoon, displaying molecules as lines, ribbons, cylinders, spheres, and other shapes. It can color molecules according to various schemes including chain, secondary structure, chemical properties, and user-defined colors.
Some key features of VMD include:
- Visualizes biomolecules using OpenGL representations such as lines, points, cylinders, spheres, and various surface models
- Can handle systems containing millions of atoms
- Calculates root mean square deviations (RMSD) between molecular structures using flexible alignment algorithms
- Interactive molecular dynamics simulation interface - can visualize and analyze results as the simulation runs
- Built-in scripting interpreter to perform advanced functions and analyses
- Extensible - new user-developed plugins can be added using Tcl and C++
VMD is developed by the Theoretical and Computational Biophysics Group at the University of Illinois at Urbana-Champaign. It is designed for molecular graphics, building molecular structures, visualizing densities from grid computing, and import/export to other formats. VMD has an extensive user community across areas like structural biology, biophysics, bioinformatics and computational chemistry.
VMD - Visual Molecular Dynamics Features
Features
- Visualization of molecular structures
- Animation of molecular dynamics trajectories
- Analysis of molecular dynamics simulations
- Support for a wide range of file formats
- Scripting and extension capabilities using Python, Tcl, or internal VMD commands
- Multi-core and GPU acceleration support
- Integrated trajectory player for animation
- Interactive molecular graphics display
Pricing
- Open Source
Pros
Cons
Official Links
Reviews & Ratings
Login to ReviewNo reviews yet
Be the first to share your experience with VMD - Visual Molecular Dynamics!
Login to ReviewThe Best VMD - Visual Molecular Dynamics Alternatives
Top Science & Education and Molecular Modeling and other similar apps like VMD - Visual Molecular Dynamics
Here are some alternatives to VMD - Visual Molecular Dynamics:
Suggest an alternative ❐PyMOL
Rasmol
Jmol
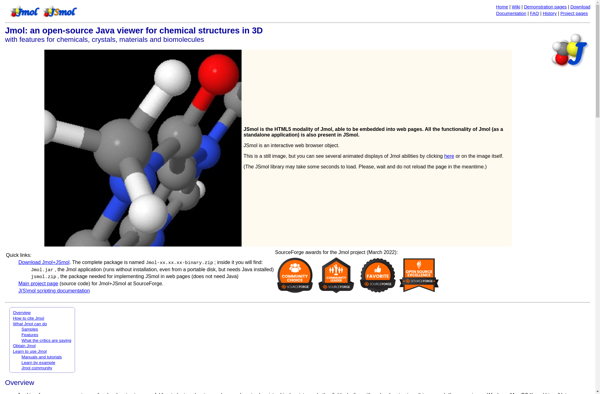
UCSF Chimera
RasTop