WinMilk
WinMilk: Biochemical Network Modeling Software
WinMilk is a free, open-source software for modeling and simulating biochemical networks. It allows users to draw pathway maps, perform simulations, and analyze experimental data. The software includes features for parameter estimation, sensitivity analysis, and much more.
What is WinMilk?
WinMilk is an open-source, Windows-based modeling and simulation software for biochemical networks and regulatory pathways. Developed at the University of Saarland in Germany, WinMilk provides researchers and students with an intuitive graphical user interface for building, analyzing, and visualizing mathematical models of biological systems.
Some key features of WinMilk include:
- User-friendly pathway editor for drawing reaction maps and setting up model equations
- Support for differential equations and Boolean networks
- Steady-state and time-course simulations using variable step size integrators
- Built-in parameter estimation tools for fitting models to experimental data
- Sensitivity analysis to identify key parameters
- Powerful scripting language for creating custom analyses
- Model import/export to SBML standard
- Production of high-quality graphs and diagrams
An advantage of WinMilk over other modeling platforms is its focus specifically on signaling/regulatory pathways rather than metabolism or cell physiology. So it includes many specialized features for studying cell signaling dynamics and transcriptional regulation.
Overall, WinMilk strikes a good balance between simplicity for beginners and advanced functionality for experts. Its wide range of analysis tools coupled with an easy-to-learn interface has made it a popular teaching tool for systems biology and bioinformatics courses.
WinMilk Features
Features
- Graphical user interface for drawing biochemical networks
- Support for various modeling frameworks like ODEs, stochastic, and Boolean
- Built-in simulations and analysis tools
- Parameter scanning and optimization
- Sensitivity analysis
- Model import/export
Pricing
- Free
- Open Source
Pros
Cons
Official Links
Reviews & Ratings
Login to ReviewThe Best WinMilk Alternatives
Top Science & Education and Biology & Chemistry and other similar apps like WinMilk
Here are some alternatives to WinMilk:
Suggest an alternative ❐Microsoft Office Outlook

Todoist
Things
ToDoList
Workflowy
Habitica
Taskade
Org mode
Task Coach
Remember The Milk
Sleek
GoTasks

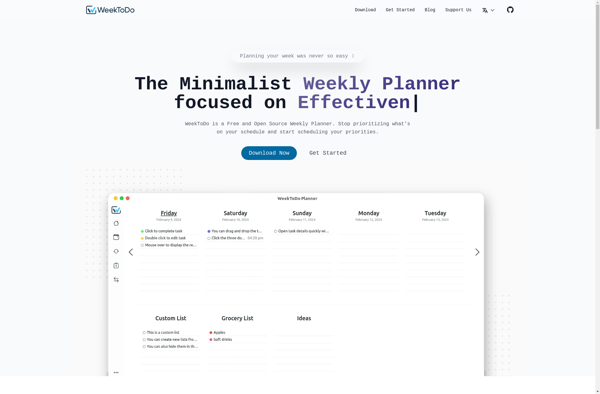
WeekToDo
OpenLoopz
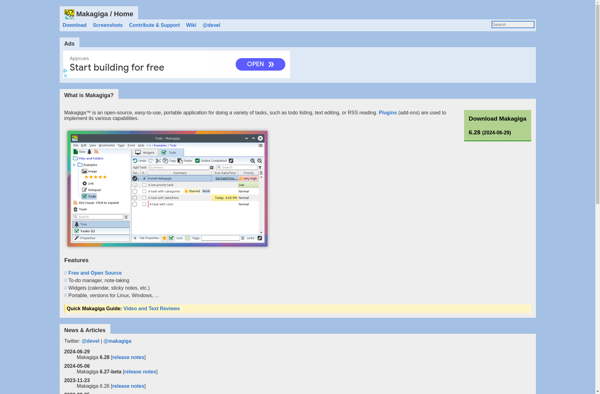
Makagiga
CloudList
ActionComplete